Our multidisciplinary lab studies HIV biology to derive insights for new therapeutic strategies.
Our current areas of focus are:
Gene Therapy, Viral Vectors, Viral Genomics, Drug Resistance, Proteomics, Host Factors, Viral Biophysics, Molecular Dynamics, HIV Entry, Hematopoiesis
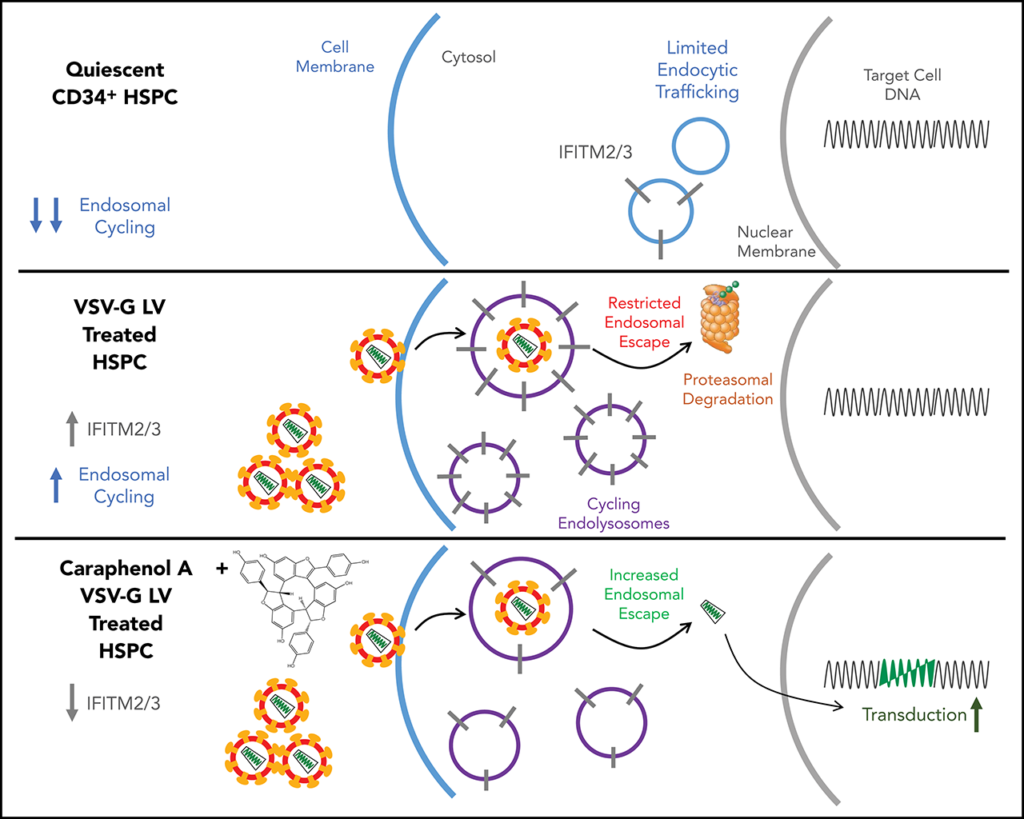
A Small Molecule Approach for Investigating Lentiviral Vector Transduction Resistance in Hematopoietic Stem Cells
Inefficient gene transfer of hematopoietic stem cells (HSCs) by lentivirus has hindered the efficient clinical application of many therapies. We are investigating various small molecules that have been found to have an effect on lentiviral transduction of HSCs and the underlying mechanism of these effects. Our goal is to improve lentiviral transduction of HSCs by better understanding and manipulating the pathways revealed from these small molecule studies.
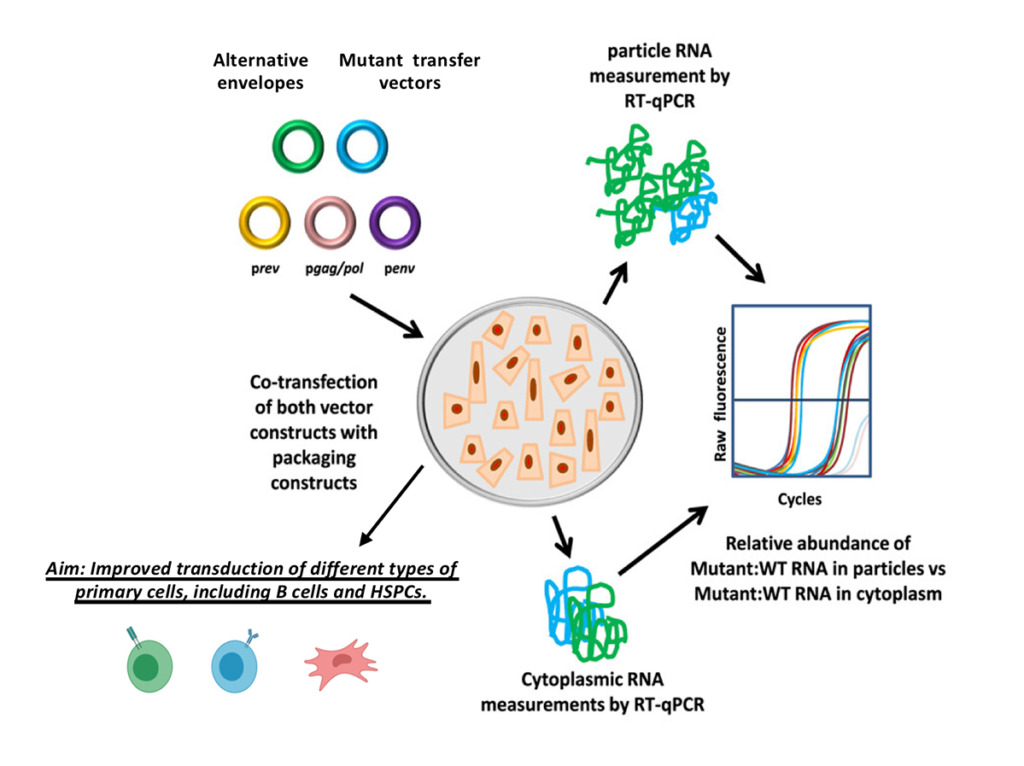
Lentiviral Vector Engineering for Improved Therapeutic Gene Delivery
Gene therapy of hematopoietic stem cells has become a promising approach for the treatment of monogenic diseases, and has been investigated as a strategy for HIV prevention and treatment. However, a large barrier to anti-HIV gene therapy remains the difficulty and inefficiency of modifying hematopoietic stem cells with current gene therapy vectors. Our lab specializes in lentiviral vector engineering using novel surface envelopes with alternate viral pseudotypes and engineering producer cell lines to increase vector titers, all with the goal of improving therapeutic gene delivery efficiency.
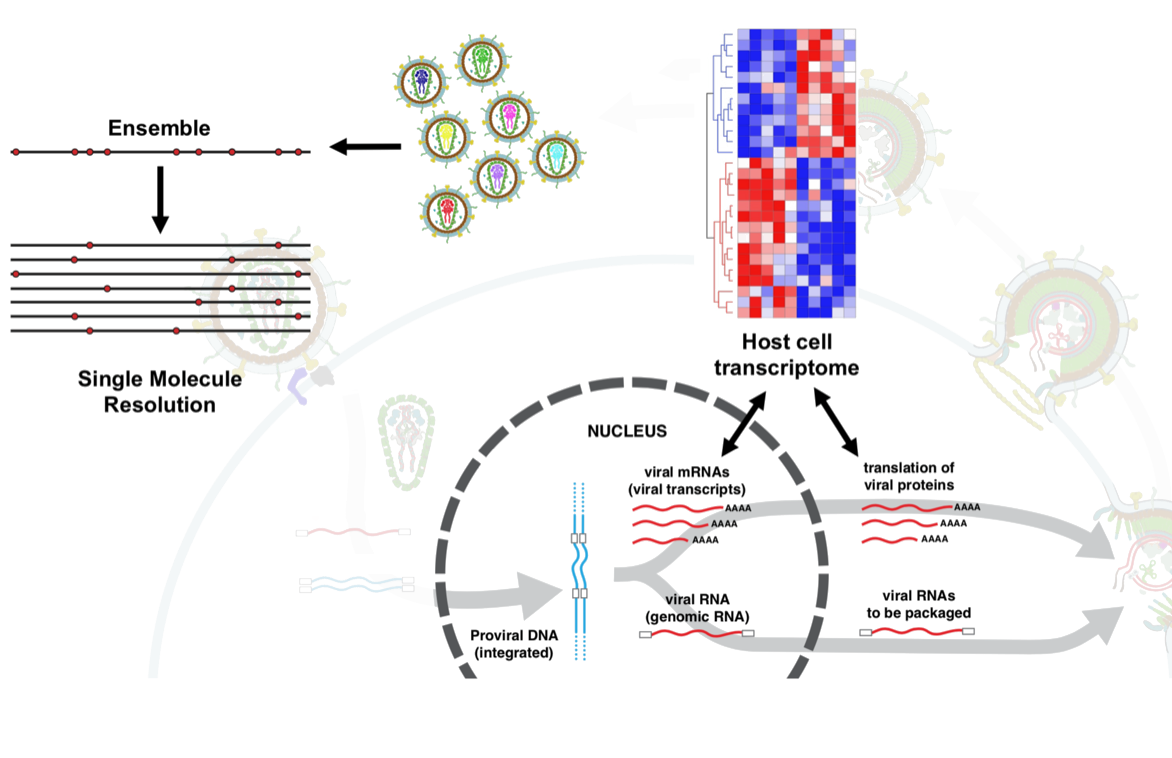
Novel sequencing strategies to interrogate in vivo viral RNA dynamics
Viral RNAs are highly dynamic and play a major role in HIV activation within host cells, and in the evolution of drug resistance to antiretroviral therapy. The development of Next-Generation Sequencing technologies has greatly contributed to understanding of the viral RNAs; however, the limited read-lengths in these platforms has curtailed the breadth of analysis of viral RNA dynamics and their biological mechanisms. To address current limitations we developed two complementary approaches for sequencing of viral mRNAs and genomic RNAs in a full-sequence context:
1) Quantitative full-length RNA-seq strategy for the simultaneous profiling of poly-adenylated HIV and host mRNA transcripts from unamplified cDNA. This approach provides granular resolution of viral and host splicing at the full isoform level allowing accurate determination of coding potential and facilitating productivity analyses.
2) Nanopore-based long-range sequencing methodology, optimized for obtaining accurate HIV cDNA sequences covering the entire Gag-Pol genomic region of circulating viruses from clinical isolates (MrHAMER). The goal is to capture novel mechanisms of drug resistance to antiretrovirals that depend on long-range functional epistatic interactions, and allow tracing of novel evolutionary pathways that result in drug failure.
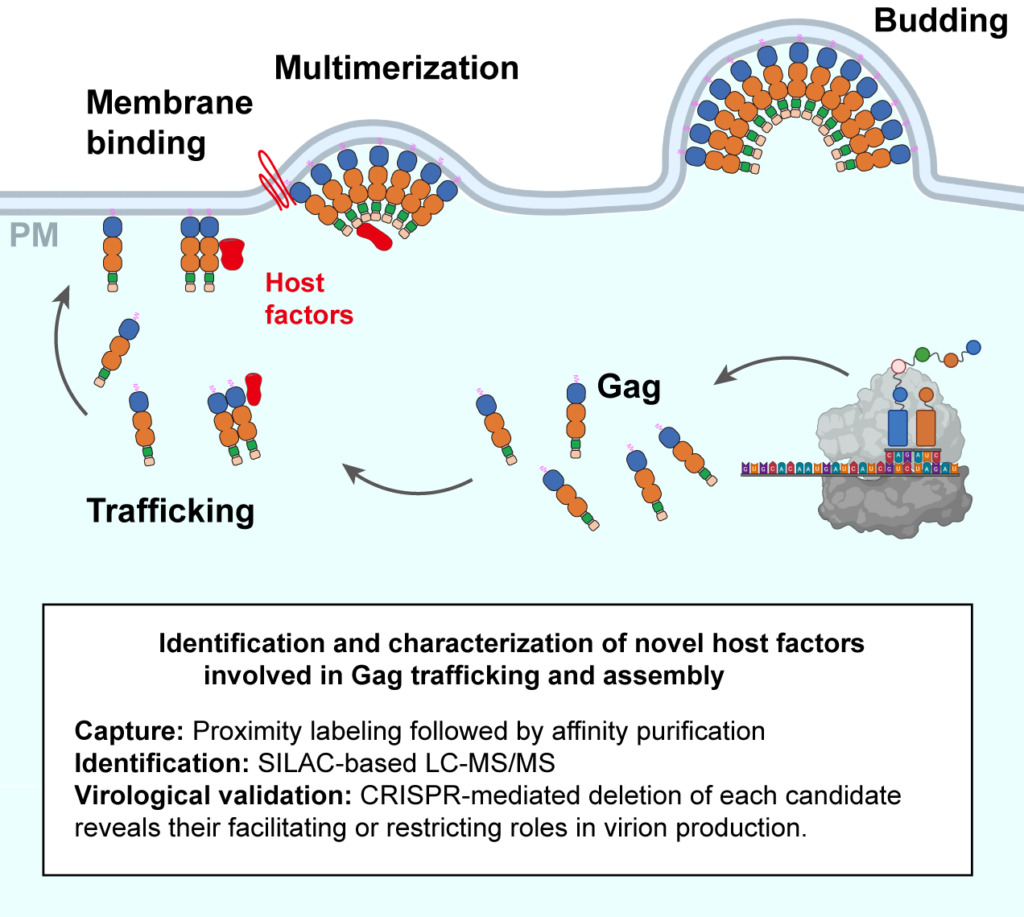
Identification and Characterization of Novel Host Factors Involved in Gag Trafficking and Assembly
Given that HIV-1 hijacks many host proteins at all stages of its life cycle, it is essential to identify these interactions as potential targets for antiretroviral therapy. HIV-1 virion formation is a multi-step process driven mainly by a structural protein called Gag. We aim to identify novel host factors associated with Gag during viral trafficking, assembly, and egress by utilizing proximity labeling and quantitative proteomic methods, then perform further biochemical and virological validations of the candidates via CRISPR and confocal imaging techniques.
Molecular mechanism for HIV transport through the Nuclear Pore Complex
In order for HIV-1 to integrate its genome into its host chromosome, the HIV-1 core must first coordinate docking, uncoating, and pre-integration complex (PIC) entry into the nucleus through the nuclear pore complex (NPC). Whereas the HIV-1 reverse transcription and integration steps are relatively well understood, the process of HIV-1 import through the nuclear pore remains poorly characterized
The Torbett lab is working towards addressing the unmet need for mechanistic and structural information on HIV-1 nuclear import, a complex process that could be targeted for novel antiviral therapy. We hypothesize that HIV-1 transport through the NPC involves multiple sequential interactions of viral components (including the capsid core) with different NUPs and novel host factors; such interactions are associated with dynamic changes in the virus and NPC structures. Specifically, we are working towards developing novel approaches to elucidate the molecular mechanisms involved in HIV-1 transport through the NPC by engineering strategies to block the inner ring of the NPC in order to study HIV-1 docking and initial entry into the NPC.
We are also engineering strategies to study the second half of transport through the NPC (HIV-1 exiting the inner ring and transport through the nuclear basket) by clogging the basket of NPC and identifying viral and host factor proteins that interact with the NPC basket during HIV-1 nuclear import.
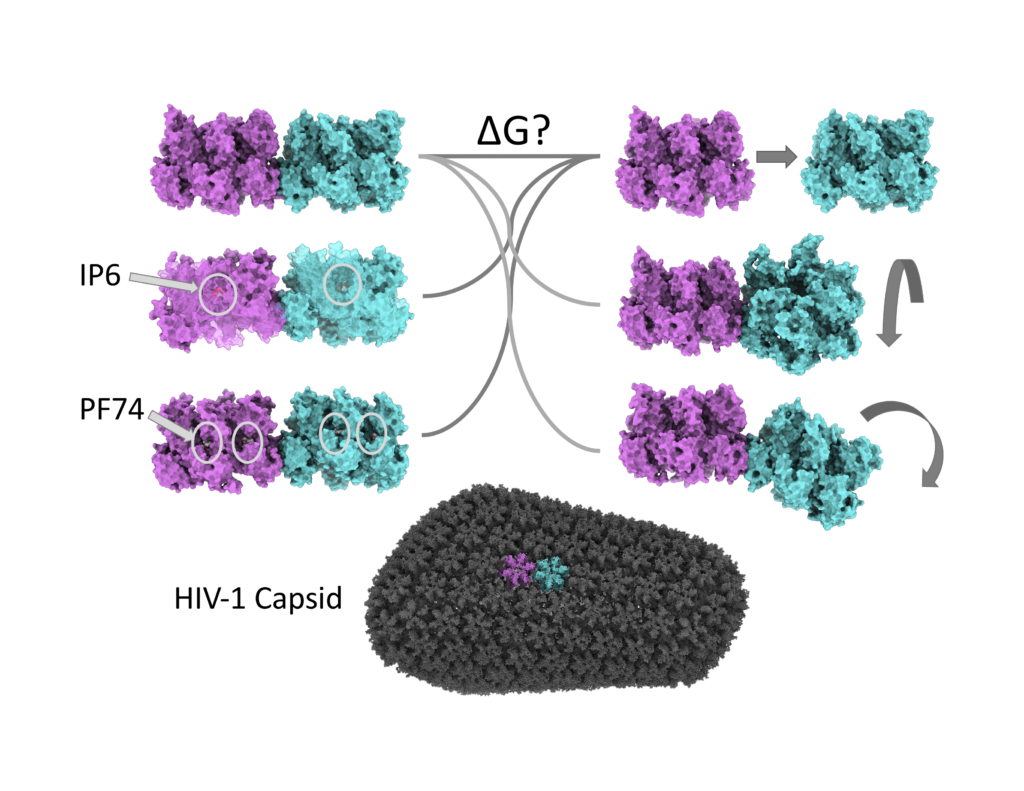
Biophysical characterization of viral capsid stability
In order to infect cells, HIV virus particles must deliver their genetic material to the cell’s nucleus. HIV is different from other viruses in that its genetic material is encased in a roughly pill-shaped container called the capsid rather than the more common spherical shell. Capsids that are too stable or unstable fail to infect the cell. It follows then that small molecules targeted at altering capsid stability are attractive, but as of yet there are no approved capsid inhibitors. The endogenous small molecule inositol hexaphosphate (IP6) and early drug candidate PF-3450074 (PF74) provide evidence for the ability of small molecules to modulate the stability of the capsid but each of their mechanisms of action are unknown. This project evaluates the impact of inherent structural variations across the capsid as well as the impact of bound small molecules on the stability of capsid protein interfaces. We will apply these findings directly to the informed design of a novel class of antiretroviral medications.
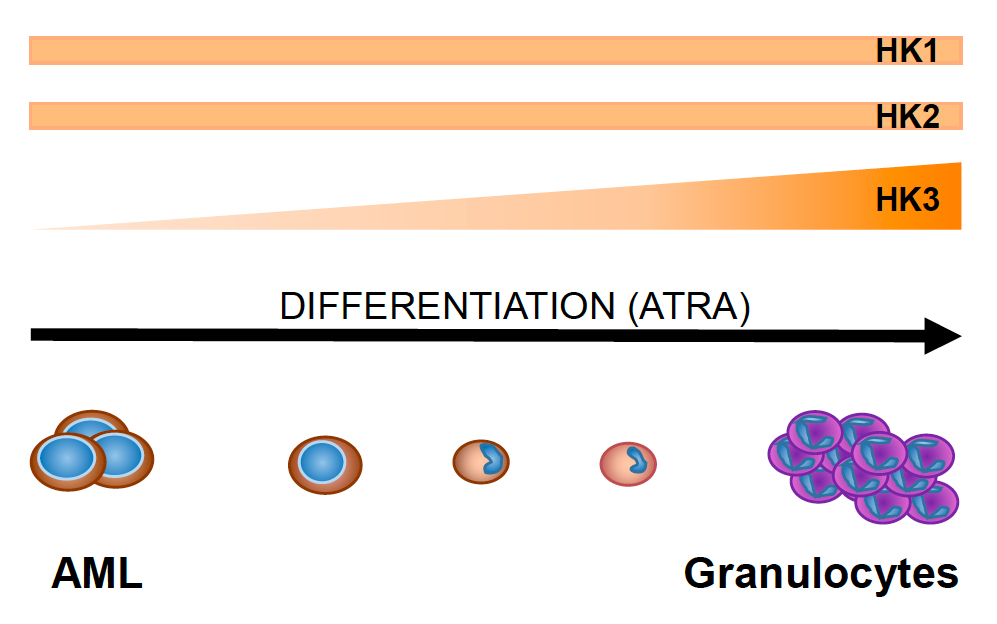
Hexokinases – Their Role in Differentiation, Metabolism and Autophagy
Hexokinases (HKs) are rate-limiting enzymes that catalyze the phosphorylation of glucose to glucose-6-phosphate, the first step of glycolysis. They promote and sustain a concentration gradient that facilitates glucose entry, which ensures the initiation of glucose dependent pathways. In general, HKs have a cytoprotective role that is highlighted by enhanced sensitivity of cancer cells to drugs when Hks were inhibited. Of the different HK isoforms, HK3 expression is significantly reduced in patient acute myeloid leukemia (AML) cells, particularly in acute promyelocytic leukemia (APL) cells expressing the PML-RARA oncofusion protein. Interestingly, while HK1 and HK2 levels remain stable during all stages of myeloid differentiation, HK3 mRNA levels significantly increase. The same pattern of HK mRNA expression was seen in NB4 APL and HL60 AML cell lines differentiated towards granulocytes and monocytes using all-trans retinoic acid (ATRA) and vitamin D3, respectively. We now aim to report about the catalytic and potential regulatory functions of HK3 during normal and aberrant myeloid hematopoiesis.